the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Numerical models for monitoring and forecasting ocean biogeochemistry: a short description of present status
Andrew Moore
Stefano Ciavatta
Katja Fennel
The ability to model biogeochemical features in the ocean is a key factor in predicting the health of the ocean: it involves the representation of processes and cycles of chemical elements (such as carbon, nutrients, and oxygen) and the dynamics of living organisms such as phytoplankton, zooplankton, and bacteria. This paper gives an overview of the main modelling aspects aimed at describing the low trophic levels of marine ecosystems and shows how they can be coupled with advection and diffusion models. The complexity of biogeochemical models can vary considerably depending on the topics of interest, assumed hypotheses, and simplifications of the numerical parameterizations. The paper also discusses the uncertainties in the numerical solution due to the lack of knowledge about the parameterizations, the initial and boundary conditions, the lack of a robust observation network, and the high computational cost of running such models.
- Article
(588 KB) - Full-text XML
- BibTeX
- EndNote
Marine biogeochemistry refers to the cycling of chemical elements (e.g. carbon, nutrients, oxygen) resulting from physical transport, chemical reactions, and uptake and processing by living organisms (e.g. phytoplankton, zooplankton, bacteria). Biogeochemical models describe the low trophic levels of marine ecosystems and are usually coupled with advection and diffusion models. Operational biogeochemical models have generally been developed by incorporating biogeochemical models developed for research and process-based studies into existing physical forecasting systems (Gehlen et al., 2015) and are used to assess (i) past and current marine ecosystem states and trends and (ii) short-term (days to weeks) or seasonal (months) forecasts (Le Traon et al., 2019; Fennel et al., 2019). When observations are assimilated, simulations of a past period are called reanalysis, while unconstrained simulations of a past period are called hindcast. When the simulations are carried forward to the present, they are referred to as nowcasts (Fennel et al., 2023). By providing timely information for the current state and a consistent reconstruction of the past, biogeochemical operational models can support ocean carbon sequestration and storage estimations, monitoring effects of acidification and deoxygenation; marine spatial planning; and, as input for habitat and food web modelling, marine biodiversity conservation and fisheries management.
Biogeochemical models can have a wide range of complexity, from a single nutrient and simple parameterizations of processes to fully explicit representations of several nutrients, trophic levels, and functional groups. They can also include carbonate systems, pollutants (e.g. Hg, persistent organic pollutants (POPs)), and other features, depending on the specific goals and domains for which they were developed. This chapter provides a brief introduction to biogeochemical modelling in the context of operational oceanography, and more detailed descriptions and discussions can be found in the following articles (Gutknecht et al., 2022; Fennel et al., 2022; Ford et al., 2018). The focus is on the levels of model complexity in existing biogeochemical prediction systems, with examples taken mainly from the Copernicus Marine Service (Le Traon et al., 2019).
2.1 Formulations, processes, and elements of biogeochemical models
In general, biogeochemical models solve a system of partial differential equations. Eq. (1) describes the rate of change of a set of state variables C representing biogeochemical tracers: dissolved inorganic substances (e.g. nutrients), living organic compartments (e.g. primary producers and secondary consumers), and non-living organic dissolved and particulate matter (Gutknecht et al., 2022; Fennel et al., 2022). The first three terms on the right-hand side of Eq. (1) represent the physical terms, advection (first term) and diffusion (second (horizontal) and third (vertical) terms) of biogeochemical tracers, where KH and KV are the horizontal and vertical diffusivities, respectively, which act on different spatial scales. The remaining terms describe the sinking processes that affect biological particles (fourth term) and biogeochemical reactions (fifth term).
The last term, Rbio, represents the local source-minus-sink terms for the biogeochemical tracers and is typically based on the principle of conservation of mass to simulate the cycling of chemical elements through various marine compartments. Biogeochemical models (Eq. 1) are generally discretized on a grid covering a spatial region of interest, and they are solved numerically by using appropriate initial and boundary conditions for each of the tracers. The physical parts of Eq. (1) can be solved directly by the advection–diffusion component of ocean dynamic models (i.e. on-line coupling). Alternatively, the output of the ocean dynamics model is used to force the biogeochemistry off-line (Heinze and Gehlen, 2013). Different schemes can be used to couple the physical and biogeochemical processes to optimize accuracy and computational cost (Bruggeman and Bolding, 2014; Cossarini et al., 2017). Operational biogeochemical models also include data assimilation schemes (Brasseur et al., 2009; Fennel et al., 2019), with satellite observations being the most commonly used due to their spatial coverage and time availability, even in near-real time. Ocean colour chlorophyll is the variable most typically assimilated in biogeochemical models (Nerger and Gregg, 2008; Ciavatta et al., 2011; Fontana et al., 2013; Teruzzi et al., 2014; Ciavatta et al., 2016), but other remote sensing variables have also been tested: diffuse attenuation coefficient (Ciavatta et al., 2014), phytoplankton functional type (PFT) chlorophyll (Ciavatta et al., 2018; Skákala et al., 2018; Pradhan et al., 2020), and inherent optical properties (Jones et al., 2016). While ocean colour provides unique information about the surface of the ocean, the transfer of surface information to deeper layers usually requires approximations. The emergence of autonomous underwater sensors (biogeochemical gliders and Argo floats) has opened the possibility to better constrain biogeochemical dynamics below the water surface (Verdy and Mazloff, 2017; Cossarini et al., 2019; Teruzzi et al., 2021; Skákala et al., 2021). Oxygen, chlorophyll, and nitrate profiles are currently used in forecast systems for assimilation (Amadio et al., 2024) but also for parameter tuning (Wang et al., 2020; Yumruktepe et al., 2023; Falls et al., 2022), validation of operational systems (Salon et al., 2019; Mignot et al., 2023), and adaptive monitoring of phytoplankton blooms (Ford et al., 2022).
Unlike physical models based on Navier–Stokes equations (Bell et al., 2025, in this report), there are no fundamental laws and principles for the biogeochemical term (Rbio). Rather, equations describing biogeochemical processes rely on empirical relationships based on laboratory experiments (e.g. nutrient limitation experiments, grazing dilution experiments), biological theories, and ecological principles based also on biogeographic relationships.
The NPZD approach, which stands for nutrient–phytoplankton–zooplankton–detritus (Fasham et al., 1990), is the basis of most marine biogeochemical models. In its simplest form, the cycling of a single nutrient (e.g. nitrogen) is represented by four marine compartments: inorganic nutrients, living organic matter (phytoplankton and zooplankton), and non-living organic matter (detritus). The nutrient fluxes between the compartments are the uptake of the nutrient as a function of phytoplankton growth, the mortality and grazing of phytoplankton and zooplankton, and the remineralization of the detritus compartment. In the original Fasham model (Fasham et al., 1990), the nutrient inorganic pool (nitrogen) is divided into ammonium and nitrate, and the remineralization process includes bacteria and dissolved organic nitrogen, increasing the number of state variables from four to seven and nearly doubling the number of processes described.
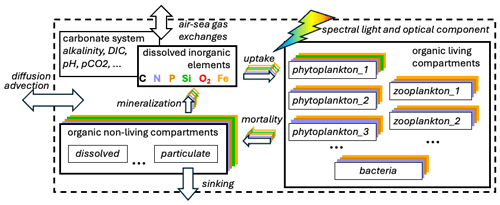
Figure 1Simplified scheme of compartments (boxes) and processes (arrows) representing the cycles of multiple chemical elements typically included in a biogeochemical model. The ellipses indicate the increase in the number of compartments and variables (e.g. multiple plankton functional types, multiple size compartments of non-living organic matter, and additional variables resolved by the carbonate system).
A schematic representation of the cycles of multiple chemical elements (e.g. nutrients) among living and non-living compartments, together with some additional features presented below, is shown in Fig. 1.
Increasing model complexity (e.g. greater number of state variables and processes) enables the expansion of the model objectives and the range of applications, but it is accompanied by larger uncertainties in the parameterization and higher computational costs. There is no general consensus on the level of complexity of biogeochemical models and the priority for new components to be added. This often depends on the specific objectives for which a model is being built. In recent years, the complexity of biogeochemical models used in operational oceanography has increased, as have their applications. These span multiple objectives: monitoring ocean state and variability, assessing ocean health (acidification, eutrophication, deoxygenation), supporting resource management, and studying pollutant impacts.
The number of chemical elements is often increased, typically including carbon, other macronutrients (such as phosphorus and silicon), and micronutrients (e.g. iron). The increase in model complexity allows modellers to represent a wider range of chemical and biological processes, such as nitrification, denitrification, calcification, competition for the limiting nutrients, and dimethylsulfide (DMS) dynamics. Another typical tracer included in biogeochemical models is oxygen because of its importance for ocean health (e.g. deoxygenation; Schmidtko et al., 2017; Grégoire et al., 2021; Bopp et al., 2013) and the effects of low oxygen concentrations (hypoxia) in changing ecosystem functions (Baird et al., 2004).
Fixed or variable nutrient stoichiometry can then be formulated within the simulated organisms, e.g. phytoplankton. Typical values of fixed nutrient ratios are 138[O2] : 106[C] : 16[N] : 15[Si] : 1[P] : 0.1–0.001[Fe] (Redfield, 1934; Lenton and Watson, 2000). When models include variable stoichiometry (e.g. Vichi et al., 2017; Tagliabue et al., 2011), multiple state variables are required to represent the living organic compartments, and a formulation of intracellular ratios can be used to simulate the multiple nutrient limitation of phytoplankton growth (Klausmeier et al., 2004). Primary production, the basis of the marine food web, is the chemical synthesis of organic compounds from dissolved carbon dioxide through chlorophyll-mediated photosynthesis. When chlorophyll is explicitly included in models, photosynthesis and acclimation to light can be dynamically simulated to balance the growth rate and the variable chlorophyll–carbon ratio as a function of light, nutrient limitation, and temperature (Geider et al., 1997).
The complexity of biogeochemical models can be measured by the number of plankton functional types (PFTs) used to simulate the trophic food web. The autotrophic community can be conceptually grouped considering various ecological functions (e.g. silicifiers, calcifiers, nitrogen fixers, and dimethylsulfide (DMS) producers); cell size (e.g. pico-, nano-, and microphytoplankton); and specific physiological traits, such as optical absorption, light use, growth rate, and affinity for nutrients (Hood et al., 2006). To improve the representation of the dynamics of phytoplankton functional groups, biogeochemical models can include a spectral radiative component which resolves solar radiation penetration in the water column (Dutkiewicz et al., 2009; Skákala et al., 2020; Álvarez et al., 2022). The zooplankton community can be subdivided by size (nano-, micro-, meso-, or macroplankton) and grazing strategy (herbivorous versus carnivorous). Additionally, a rigid partition between autotrophs and heterotrophs is not exhaustive, and the food web can incorporate mixotrophs to account for organisms that obtain energy through both photosynthesis and consumption of others (Flynn et al., 2013; Mitra et al., 2014).
Biogeochemical models can target biodiversity (Litchman and Klausmeier, 2008) if the number of functional plankton groups is large enough to deterministically represent niches based on certain factors (e.g. adaptation to the light spectrum; Álvarez et al., 2022) or by including tens or hundreds of PFTs with randomly prescribed parameters so that the fittest groups can prevail in the resulting ecosystem (Follows et al., 2007).
Assessing ocean carbon uptake and the associated ocean acidification requires modelling of the marine carbonate system: the two prognostic variables are typically dissolved inorganic carbon and alkalinity, and carbonate chemistry is solved to determine water acidity and to calculate the air–sea CO2 gas exchange (Zeebe and Wolf-Gladrow, 2001; Artioli et al., 2012; Cossarini et al., 2015a).
The microbial loop describes the role of bacteria in decomposing organic matter that is converted back to nutrients. It also includes the channelling of energy and matter to higher trophic levels (HTLs) by microzooplankton, which can be an important pathway in oligotrophic conditions (Legendre and Rassoulzadegan, 1995; Hood et al., 2006). In addition, models can describe the dynamics of multiple pools of dissolved organic matter (e.g. labile, semilabile, and refractory) characterized by turnover timescales ranging from days to years (Anderson et al., 2015; Glibert and Mitra, 2022). In coastal- and shallow-water applications, a benthic model allows us to represent the mutual interaction and nutrient, carbon, and oxygen exchanges between the water column and the sediment (Soetaert et al., 2000).
Biogeochemical models can be linked or coupled to higher-trophic-level or ecosystem models (Libralato, 2025, in this report). This requires parameterization or explicit representation of the link of phytoplankton productivity and zooplankton mortality with higher-trophic-level (HTL) compartments, such as nekton and fishes, and possibly the feedbacks from HTLs to biogeochemical processes (e.g. Travers et al., 2009).
Although not yet ready to be integrated into an operational prediction system, additional – and useful to society – components of biogeochemical models could include the fate, dynamics, and transport of metals and POPs (Melaku Canu et al., 2015; Wagner et al., 2019, Bieser et al., 2023), including bioaccumulation in low-trophic-level compartments and cumulative impacts on marine species and ecosystems (Rosati et al., 2022; Zhang et al., 2020).
2.2 Main models used in operational systems
Unlike ocean dynamics, where a limited number of numerical models are used in operational applications (e.g. NEMO, ROMS, MITGCM; see Alvarez-Fanjul et al., 2022), there is a long list of biogeochemical models that have varying levels of complexity in response to specific regions and topics of interest for which they were developed. As part of the UN Decade of Ocean Science for Sustainable Development programme, the Decade Collaborative Centre for Ocean Prediction (Alvarez-Fanjul et al., 2024) is promoting the Atlas of Operational Systems, which also describes their biogeochemical component (https://www.unoceanprediction.org/en/homepage, last access: 18 April 2025). Some of the biogeochemical models used in operational prediction systems are briefly presented below, roughly ordered by increasing complexity:
-
HadOCC (Palmer and Totterdell, 2001). A model of low complexity (10 variables) with a single phytoplankton and single zooplankton, with fixed stoichiometry used to produce global reanalysis of the carbon cycle (Ford and Barciela, 2017).
-
SCOBI. Used for reanalysis of nutrient cycling in the Baltic Sea (Liu et al., 2017), it has fixed nutrient stoichiometry in three phytoplankton and one zooplankton and includes anaerobic processes and a sediment module for oxygen and nutrient dynamics (Eilola et al., 2009).
-
NEMURO (Kishi et al., 2007). A relatively simple low-trophic-level model of the Pacific Ocean (11 state variables), based on N dynamics with two phytoplankton and two zooplankton, that has been coupled with an HLT model (e.g. bioenergetic fish model; Kishi et al., 2011).
-
ECB (Feng et al., 2015). Developed to study eutrophication in the Chesapeake Bay, it consists of 11 variables (C and N cycles) with one single phytoplankton and one single zooplankton and processes applicable for estuarine ecosystems, such as inorganic suspended solid dynamics and the impact on light attenuation (Feng et al., 2015; Irby and Friedrichs, 2019; Irby et al., 2018).
-
GulfMexico. Developed to investigate eutrophication and acidification in the Gulf of Mexico, it is a model of intermediate complexity (15 variables) that simulates N, P, O2, and C dynamics with a single phytoplankton and single zooplankton group, a sediment-water flux parameterization, and the carbonate system (Fennel et al., 2011; Laurent et al., 2017).
-
PISCES. A model of intermediate complexity (24 state variables) with five nutrients, fixed stoichiometry, and two phytoplankton and two zooplankton size classes, it includes carbonate system and dissolved oxygen dynamics (Aumont et al., 2015). It is currently used in regional (northeastern Atlantic; Gutknecht et al., 2019) and global operational systems (Mignot et al., 2023). A version with variable stoichiometry (PISCES-QUOTA) also exists and is used for climate scenario studies (Kwiatkowski et al., 2018).
-
ECOSMO (Daewel and Schrum, 2013). Its operational version in use for the Northern Atlantic and Arctic oceans (Yumruktepe et al., 2022) has two phytoplankton, two zooplankton, multiple nutrients (N, P, and Si), and a fixed molar Redfield ratio but variable chlorophyll-to-carbon dynamics, and it includes a nutrient sediment layer.
-
ERGOM. Used in the Baltic Sea operational system, it is a model of intermediate complexity (25 variables) with three phytoplankton and two zooplankton groups, and it includes processes related to hypoxia and anoxia, a carbonate system, and a radiative model with dynamics for coloured dissolved organic matter (Neumann, 2000; Neumann et al., 2015).
-
BAMHBI. Developed for the Black Sea, which is characterized by an anoxic deep layer, the model includes 33 pelagic state variables, with multiple nutrients and eight plankton functional types, and explicitly describes processes in the anoxic layer. It also includes dynamics of the sedimentary stocks of organic C, N, P, and biogenic Si (Grégoire et al., 2008; Grégoire and Soetaert, 2010; Capet et al., 2016; Ciliberti et al., 2022)
-
eReefs/vB3p0. Designed for water quality in the Australian Great Barrier Reef marine ecosystem, it is a complex model resolving N, P, C, and O2 cycles in pelagic (four phytoplankton and two zooplankton) and sediment (seagrass and coral) environments. It includes carbonate chemistry (Mongin et al., 2016), bio-optics, and bleaching (Baird et al., 2016, 2020).
-
BFM. A multi-nutrient and multi-plankton model (Álvarez et al., 2022) with more than 50 variables, it includes carbonate chemistry (Cossarini et al., 2015b, 2017), bio-optics (Lazzari et al., 2021), and pollutants (Rosati et al., 2022) and is currently used in the operational system and reanalysis of the Mediterranean Sea (Salon et al., 2019; Cossarini et al., 2021).
-
ERSEM. Developed for regional (northeastern Atlantic and North Sea) and global studies, it is a complex model (more than 50 state variables) including multiple nutrients, multi-plankton, a carbonate system, and a sediment layer (Baretta et al., 1995; Butenschön et al., 2016).
-
DARWIN. A complex multi-nutrient model in which the plankton community comprises hundreds of groups by taking into account cell size and functional traits to study the biodiversity and biogeography (Dutkiewicz et al., 2009).
Given the complexity of marine ecosystems, the development of biogeochemical models is the result of compromises and simplifications, and no single approach can realistically encompass all relevant aspects of marine ecosystem dynamics. Determining the appropriate level of complexity depends on the specific objectives and supporting information for each application, while standard assessment frameworks (Hernandez et al., 2018) represent essential tools to assess model performance. Increasing model complexity does not necessarily mean better performance (Xiao and Friedrichs, 2014; Kwiatkowski et al., 2014; Gehlen et al., 2015; Séférian et al., 2020). Indeed, despite recent significant technological advances in observing systems, the lack of biogeochemical observations, both in terms of the number of variables and spatiotemporal availability, remains the major obstacle for thorough validation and optimization (e.g. tuning parameters).
In addition to inherent uncertainties in model structures and parameterizations, important sources of uncertainty arise from numerical solution settings in spatially discretized domains, e.g. initial conditions, lateral conditions for open boundaries, inputs of chemical compounds and suspended matter from rivers and atmospheric deposition, and ocean dynamics driving the transport of biogeochemical tracers. Despite major advances in high-performance computing, the computational cost of a model still constitutes an issue when the resolution of the numerical solution and the complexity (e.g. the number of biogeochemical tracers) are increased. Many of the biogeochemical models have emerged as community models that should guarantee a distributed and affordable effort to keep them up to date with the advancement in marine ecology knowledge and the requirements of evolving computer science and of the coupling with physical and Earth system models and data assimilation frameworks. Rapidly evolving applications of artificial intelligence in marine biogeochemistry can assist in optimizing model parameters, developing hybrid models to improve predictions and operational system efficiency, and detecting patterns in large data sets from reanalysis. Linking microbial community dynamics to ecosystem processes through metagenomic data can improve models describing nutrient cycling, carbon fluxes, and diversity. New coupling paradigms are needed to promote the integration of biogeochemical models with the dynamics of pollutants, high trophic levels, and Earth system components.
In addition to science-driven developments, operational biogeochemical systems can evolve to respond to societal demands to assess the impacts of heat waves, oxygen depletion and acidification on marine resources, and the role of the oceans in achieving the goal of carbon neutrality.
No data sets were used in this article.
GC prepared the first draft, and all co-authors participated in the review and finalization.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
The authors thank the three referees for their thoughtful and constructive feedback.
This paper was edited by Stefania Angela Ciliberti and reviewed by three anonymous referees.
Álvarez, E., Lazzari, P., and Cossarini, G.: Phytoplankton diversity emerging from chromatic adaptation and competition for light, Prog. Oceanogr., 204, 102789, https://doi.org/10.1016/j.pocean.2022.102789, 2022.
Alvarez-Fanjul, E., Ciliberti, S., and Bahurel, P.: Implementing Operational Ocean Monitoring and Forecasting Systems. IOC-UNESCO, GOOS-275, https://doi.org/10.48670/ETOOFS, 2022.
Alvarez-Fanjul, E., Ciliberti, S., Pearlman, J., Wilmer-Becker, K., Bahurel, P., Ardhuin, F., Arnaud, A., Azizzadenesheli, K., Aznar, R., Bell, M., Bertino, L., Behera, S., Brassington, G., Calewaert, J. B., Capet, A., Chassignet, E., Ciavatta, S., Cirano, M., Clementi, E., Cornacchia, L., Cossarini, G., Coro, G., Corney, S., Davidson, F., Drevillon, M., Drillet, Y., Dussurget, R., El Serafy, G., Fearon, G., Fennel, K., Ford, D., Le Galloudec, O., Huang, X., Lellouche, J. M., Heimbach, P., Hernandez, F., Hogan, P., Hoteit, I., Joseph, S., Josey, S., Le Traon, P.-Y., Libralato, S., Mancini, M., Martin, M., Matte, P., McConnell, T., Melet, A., Miyazawa, Y., Moore, A. M., Novellino, A., O'Donncha, F., Porter, A., Qiao, F., Regan, H., Robert-Jones, J., Sanikommu, S., Schiller, A., Siddorn, J., Sotillo, M. G., Staneva, J., Thomas-Courcoux, C., Thupaki, P., Tonani, M., Garcia Valdecasas, J. M., Veitch, J., von Schuckmann, K., Wan, L., Wilkin, J., Zhong, A., and Zufic, R.: Promoting best practices in ocean forecasting through an Operational Readiness Level, Front. Mar. Sci., 11, 1443284, https://doi.org/10.3389/fmars.2024.1443284, 2024.
Amadio, C., Teruzzi, A., Pietropolli, G., Manzoni, L., Coidessa, G., and Cossarini, G.: Combining neural networks and data assimilation to enhance the spatial impact of Argo floats in the Copernicus Mediterranean biogeochemical model, Ocean Sci., 20, 689–710, https://doi.org/10.5194/os-20-689-2024, 2024.
Anderson, T. R., Christian, J. R., and Flynn, K. J.: Chapter 15 – Modeling DOM Biogeochemistry, Biogeochemistry of Marine Dissolved Organic Matter (Second Edition), edited by: Hansell, D. A. and Carlson, C. A., Academic Press, 635–667, https://doi.org/10.1016/B978-0-12-405940-5.00015-7, 2015.
Artioli, Y., Blackford, J. C., Butenschön, M., Holt, J. T., Wakelin, S. L., Thomas, H., Borges, A. V., and Allen, J. I.: The carbonate system in the North Sea: Sensitivity and model validation, J. Marine Syst., 102, 1–13, https://doi.org/10.1016/j.jmarsys.2012.04.006, 2012.
Aumont, O., Ethé, C., Tagliabue, A., Bopp, L., and Gehlen, M.: PISCES-v2: an ocean biogeochemical model for carbon and ecosystem studies, Geosci. Model Dev., 8, 2465–2513, https://doi.org/10.5194/gmd-8-2465-2015, 2015.
Baird, D., Christian, R. R., Peterson, C. H., and Johnson, G. A.: Consequences of hypoxia on estuarine ecosystem function: energy diversion from consumers to microbes, Ecol. Appl., 14, 805–822, https://doi.org/10.1890/02-5094, 2004.
Baird, M. E., Cherukuru, N., Jones, E., Margvelashvili, N., Mongin, M., Oubelkheir, K., and Wild-Allen, K. A.: Remote-sensing reflectance and true colour produced by a coupled hydrodynamic, optical, sediment, biogeochemical model of the Great Barrier Reef, Australia: comparison with satellite data. Environ. Model. Softw., 78, 79–96, https://doi.org/10.1016/j.envsoft.2015.11.025, 2016.
Baird, M. E., Wild-Allen, K. A., Parslow, J., Mongin, M., Robson, B., Skerratt, J., Rizwi, F., Soja-Woźniak, M., Jones, E., Herzfeld, M., Margvelashvili, N., Andrewartha, J., Langlais, C., Adams, M. P., Cherukuru, N., Gustafsson, M., Hadley, S., Ralph, P. J., Rosebrock, U., Schroeder, T., Laiolo, L., Harrison, D., and Steven, A. D. L.: CSIRO Environmental Modelling Suite (EMS): scientific description of the optical and biogeochemical models (vB3p0), Geosci. Model Dev., 13, 4503–4553, https://doi.org/10.5194/gmd-13-4503-2020, 2020.
Baretta, J. W., Ebenhöh, W., and Ruardij, P.: The European regional seas ecosystem model, a complex marine ecosystem model, Neth. J. Sea Res., 33, 233–246, https://doi.org/10.1016/0077-7579(95)90047-0, 1995.
Bell, M. J., Schiller, A., and Ciliberti, S.: Numerical Models for Simulating Ocean Physics, in: Ocean prediction: present status and state of the art (OPSR), edited by: Álvarez Fanjul, E., Ciliberti, S. A., Pearlman, J., Wilmer-Becker, K., and Behera, S., Copernicus Publications, State Planet, 5-opsr, 10, https://doi.org/10.5194/sp-5-opsr-10-2025, 2025.
Bieser, J., Amptmeijer, D. J., Daewel, U., Kuss, J., Soerensen, A. L., and Schrum, C.: The 3D biogeochemical marine mercury cycling model MERCY v2.0 – linking atmospheric Hg to methylmercury in fish, Geosci. Model Dev., 16, 2649–2688, https://doi.org/10.5194/gmd-16-2649-2023, 2023.
Bopp, L., Resplandy, L., Orr, J. C., Doney, S. C., Dunne, J. P., Gehlen, M., Halloran, P., Heinze, C., Ilyina, T., Séférian, R., Tjiputra, J., and Vichi, M.: Multiple stressors of ocean ecosystems in the 21st century: projections with CMIP5 models, Biogeosciences, 10, 6225–6245, https://doi.org/10.5194/bg-10-6225-2013, 2013.
Brasseur, P., Gruber, N., Barciela, R., Brander, K., Doron, M., El Moussaoui, A., Hobday, A.J., Huret, M., Kremeur, A.-S., Lehodey, P., Matear, R., Moulin, C., Murtugudde, R., Senina, I., and Svendsen, E.: Integrating biogeochemistry and ecology into ocean data assimilation systems, Oceanography, 22, 206–215, https://doi.org/10.5670/oceanog.2009.80, 2009.
Bruggeman, J. and Bolding, K.: A general framework for aquatic biogeochemical models, Environ. Model. Softw., 61, 249–265, https://doi.org/10.1016/j.envsoft.2014.04.002, 2014.
Butenschön, M., Clark, J., Aldridge, J. N., Allen, J. I., Artioli, Y., Blackford, J., Bruggeman, J., Cazenave, P., Ciavatta, S., Kay, S., Lessin, G., van Leeuwen, S., van der Molen, J., de Mora, L., Polimene, L., Sailley, S., Stephens, N., and Torres, R.: ERSEM 15.06: a generic model for marine biogeochemistry and the ecosystem dynamics of the lower trophic levels, Geosci. Model Dev., 9, 1293–1339, https://doi.org/10.5194/gmd-9-1293-2016, 2016.
Capet, A., Meysman, F. J. R., Akoumianaki, I., Soetaert, K., and Grégoire, M.: Integrating sediment biogeochemistry into 3D oceanic models: A study of benthic-pelagic coupling in the Black Sea, Ocean Model., 101, 83–100, https://doi.org/10.1016/j.ocemod.2016.03.006, 2016.
Ciavatta, S., Torres, R., Saux-Picart, S., and Allen, J. I.: Can ocean color assimilation improve biogeochemical hindcasts in shelf seas?, J. Geophys. Res.-Oceans, 116, C12043, https://doi.org/10.1029/2011JC007219, 2011.
Ciavatta, S., Torres, R., Martinez-Vicente, V., Smyth, T., Dall'Olmo, G., Polimene, L., and Allen, J. I.: Assimilation of remotely-sensed optical properties to improve marine biogeochemistry modelling, Prog. Oceanogr., 127, 74–95, 2014.
Ciavatta, S., Kay, S., Saux-Picard, S., Butenschön, M., and Allen, J. I.: Decadal reanalysis of biogeochemical indicators and fluxes in the North West European shelf-sea ecosystem, J. Geophys. Res.-Oceans, 121, 1824–1845, https://doi.org/10.1002/2015JC011496, 2016.
Ciavatta, S., Brewin, R. J. W., Skákala, J., Polimene, L., de Mora, L., Artioli, Y., and Allen, J. I.: Assimilation of ocean-color plankton functional types to improve marine ecosystem simulations, J. Geophys. Res.-Oceans, 123, 834–854, 2018.
Ciliberti, S. A., Grégoire, M., Staneva, J., Palazov, A., Coppini, G., Lecci, R., Peneva, E., Matreata, M., Marinova, V., Masina, S., and Pinardi, N.: Monitoring and Forecasting the Ocean State and Biogeochemical Processes in the Black Sea: Recent Developments in the Copernicus Marine Service, J. Mar. Sci. Eng., 9, 1146, https://doi.org/10.3390/jmse9101146, 2022.
Cossarini, G., Querin, S., and Solidoro, C.: The continental shelf carbon pump in the northern Adriatic Sea (Mediterranean Sea): Influence of wintertime variability, Ecol. Model., 314, 118–134, https://doi.org/10.1016/j.ecolmodel.2015.07.024, 2015a.
Cossarini, G., Lazzari, P., and Solidoro, C.: Spatiotemporal variability of alkalinity in the Mediterranean Sea, Biogeosciences, 12, 1647–1658, https://doi.org/10.5194/bg-12-1647-2015, 2015b.
Cossarini, G., Querin, S., Solidoro, C., Sannino, G., Lazzari, P., Di Biagio, V., and Bolzon, G.: Development of BFMCOUPLER (v1.0), the coupling scheme that links the MITgcm and BFM models for ocean biogeochemistry simulations, Geosci. Model Dev., 10, 1423–1445, https://doi.org/10.5194/gmd-10-1423-2017, 2017.
Cossarini, G., Mariotti, L., Feudale, L., Mignot, A., Salon, S., Taillandier, V., Teruzzi, A., and D'Ortenzio, F.: Towards operational 3D-Var assimilation of chlorophyll Biogeochemical-Argo float data into a biogeochemical model of the Mediterranean Sea, Ocean Model., 133, 112–128, https://doi.org/10.1016/j.ocemod.2018.11.005, 2019.
Cossarini G., Feudale, L., Teruzzi, A., Bolzon, G., Coidessa, G., Solidoro, C., Di Biagio, V., Amadio, C., Lazzari, P., Brosich, A., and Salon, S.: High-Resolution Reanalysis of the Mediterranean Sea Biogeochemistry (1999–2019), Front. Mar. Sci., 8, 1–21, https://doi.org/10.3389/fmars.2021.741486, 2021.
Daewel, U. and Schrum, C.: Simulating long-term dynamics of the coupled North Sea and Baltic Sea ecosystem with ECOSMO II: Model description and validation. J. Marine Syst., 119–120, 30–49, https://doi.org/10.1016/j.jmarsys.2013.03.008, 2013.
Dutkiewicz, S., Follows, M. J., and Bragg, J. G.: Modeling the coupling of ocean ecology and biogeochemistry, Global Biogeochem. Cy., 23, 1–15, 2009.
Eilola, K., Meier, H. E. M., and Almroth, E.: On the dynamics of oxygen, phosphorus and cyanobacteria in the Baltic Sea: a model study, J. Marine Syst., 75, 163–184, https://doi.org/10.1016/j.jmarsys.2008.08.009, 2009.
Falls, M., Bernardello, R., Castrillo, M., Acosta, M., Llort, J., and Galí, M.: Use of genetic algorithms for ocean model parameter optimisation: a case study using PISCES-v2_RC for North Atlantic particulate organic carbon, Geosci. Model Dev., 15, 5713–5737, https://doi.org/10.5194/gmd-15-5713-2022, 2022.
Fasham, M. J. R., Ducklow, H. W., and McKelvie, S. M.: A nitrogen-based model of plankton dynamics in the oceanic mixed layer, J. Mar. Res., 48, 591–639, https://elischolar.library.yale.edu/journal_of_marine_research/1981 (last access: 18 April 2025), 1990.
Feng, Y., Friedrichs, M. A. M., Wilkin, J., Tian, H., Yang, Q., Hofmann, E. E., Wiggert, J. D., and Hood, R. R.: Chesapeake Bay nitrogen fluxes derived from a land-estuarine-ocean biogeochemical modeling system: model description, evaluation and nitrogen budgets, J. Geophys. Res.-Biogeo., 120, 1666–1695, https://doi.org/10.1002/2015JG002931, 2015.
Fennel, K., Hetland, R., Feng, Y., and DiMarco, S.: A coupled physical-biological model of the Northern Gulf of Mexico shelf: model description, validation and analysis of phytoplankton variability, Biogeosciences, 8, 1881–1899, https://doi.org/10.5194/bg-8-1881-2011, 2011.
Fennel, K., Gehlen, M., Brasseur, P., Brown, C. W., Ciavatta, S., Cossarini, G., Crise, A., Edwards, C. A., Ford, D., Friedrichs, M. A., and Gregoire, M.: Advancing marine biogeochemical and ecosystem reanalyses and forecasts as tools for monitoring and managing ecosystem health, Front. Mar. Sci., 6, 89, https://doi.org/10.3389/fmars.2019.00089, 2019.
Fennel, K., Mattern, J. P., Doney, S. C., Bopp, L., Moore, A. M., Wang, B., and Yu, L.: Ocean biogeochemical modelling, Nature Reviews Methods Primers, 2, 76, https://doi.org/10.1038/s43586-022-00154-2, 2022.
Fennel, K., Long, M. C., Algar, C., Carter, B., Keller, D., Laurent, A., Mattern, J. P., Musgrave, R., Oschlies, A., Ostiguy, J., Palter, J. B., and Whitt, D. B.: Modelling considerations for research on ocean alkalinity enhancement (OAE), in: Guide to Best Practices in Ocean Alkalinity Enhancement Research, edited by: Oschlies, A., Stevenson, A., Bach, L. T., Fennel, K., Rickaby, R. E. M., Satterfield, T., Webb, R., and Gattuso, J.-P., Copernicus Publications, State Planet, 2-oae2023, 9, https://doi.org/10.5194/sp-2-oae2023-9-2023, 2023.
Flynn, K. J., Stoecker, D. K., Mitra, A., Raven, J. A., Glibert, P. M., Hansen, P. J., Granéli, E., and Burkholder, J. M.: Misuse of the phytoplankton–zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types, J. Plankton Res., 35, 3–11, https://doi.org/10.1093/plankt/fbs062, 2013.
Follows, M. J., Dutkiewicz, S. Grant, S., and Chisholm, S.W.: Emergent biogeography of microbial communities in a model ocean, Science, 315, 1843–1846, https://doi.org/10.1126/science.1138544, 2007.
Fontana, C., Brasseur, P., and Brankart, J.-M.: Toward a multivariate reanalysis of the North Atlantic Ocean biogeochemistry during 1998–2006 based on the assimilation of SeaWiFS chlorophyll data, Ocean Sci., 9, 37–56, https://doi.org/10.5194/os-9-37-2013, 2013.
Ford, D. and Barciela, R.: Global marine biogeochemical reanalyses assimilating two different sets ofmerged ocean colour products, Remote Sens. Environ., 203, 40–54, https://doi.org/10.1016/j.rse.2017.03.040, 2017.
Ford, D., Key, S., McEwan, R., Totterdell, I., and Gehlen, M.: Marine biogeochemical modelling and data assimilation for operational forecasting, reanalysis, and climate research, in: New Frontiers in Operational Oceanography, edited by: Chassignet, E., Pascual, A., Tintoré, J., and Verron, J., GODAE OceanView, 625–652, https://doi.org/10.17125/gov2018.ch22, 2018.
Ford, D. A., Grossberg, S., Rinaldi, G., Menon, P. P., Palmer, M. R., Skakala, J., Smyth, T., Williams, C. A. J., Lopez, A. L., and Ciavatta, S.: A solution for autonomous, adaptive monitoring of coastal ocean ecosystems: Integrating ocean robots and operational forecasts, Front. Mar. Sci., 9, 1067174, https://doi.org/10.3389/fmars.2022.1067174, 2022.
Gehlen, M., Barciela, R., Bertino, L., Brasseur, P., Butenschön, M., Chai, F., and Simon, E.: Building the capacity for forecasting marine biogeochemistry and ecosystems: recent advances and future developments, J. Oper. Oceanogr., 8, s168–s187, https://doi.org/10.1080/1755876X.2015.1022350, 2015.
Geider, R. J., MacIntyre, H. L., and Kana, T. M.: Dynamic model of phytoplankton growth and acclimation: responses of the balanced growth rate and the chlorophyll a: carbon ratio to light, nutrient-limitation and temperature, Mar. Ecol. Prog. Ser., 148, 187–200, http://www.jstor.org/stale/24857483 (last access: 18 April 2025), 1997.
Glibert, P. M. and Mitra, A.: From webs, loops, shunts, and pumps to microbial multitasking: Evolving concepts of marine microbial ecology, the mixoplankton paradigm, and implications for a future ocean, Limnol. Oceanogr., 67, 585–597, https://doi.org/10.1002/lno.12018, 2022.
Grégoire, M. and Soetaert, K.: Carbon, nitrogen, oxygen and sulfide budgets in the Black Sea: A biogeochemical model of the whole water column coupling the oxic and anoxic parts, Ecol. Model., 221, 2287–2301, https://doi.org/10.1016/j.ecolmodel.2010.06.007, 2010.
Grégoire, M., Raick, C., and Soetaert, K.: Numerical modeling of the deep Black Sea ecosystem functioning during the late 80's (eutrophication phase), Prog. Oceanogr., 76, 286–333, https://doi.org/10.1016/j.pocean.2008.01.002, 2008.
Grégoire, M., Garçon, V., Garcia, H., Breitburg, D., Isensee, K., Oschlies, A., Telszewski, M., Barth, A., Bittig, H.C., Carstensen, J., and Carval, T.: A global ocean oxygen database and atlas for assessing and predicting deoxygenation and ocean health in the open and coastal ocean. Front. Mar. Sci., 8, 724913, https://doi.org/10.3389/fmars.2021.724913, 2021.
Gutknecht, E., Reffray, G., Mignot, A., Dabrowski, T., and Sotillo, M. G.: Modelling the marine ecosystem of Iberia–Biscay–Ireland (IBI) European waters for CMEMS operational applications, Ocean Sci., 15, 1489–1516, https://doi.org/10.5194/os-15-1489-2019, 2019.
Gutknecht, E., Bertino, L., Brasseur, P., Ciavatta, S., Cossarini, G., Fennel, K., Ford, D., Grégoire M., Lavoie D., and Lehodey, P.: Biogeochemical Modelling, in: Implementing Operational Ocean Monitoring and Forecasting Systems, edited by: Alvarez-Franjul, E., Ciliberti, S., and Bahurel, P., IOC-UNESCO, GOOS-275, https://doi.org/10.48670/ETOOFS, 2022.
Heinze, C. and Gehlen, M.: Chapter 26: Modelling ocean biogeochemical processes and resulting tracer distributions, Int. Geophys., 103, 667–694, https://doi.org/10.1016/B978-0-12-391851-2.00026-X, 2013.
Hernandez, F., Smith, G., Baetens, K., Cossarini, G., Garcia-Hermosa, I., Drévillon, M., Maksymczuk, J., Melet, A., Régnier, C., and Schuckmann, K. V.: Measuring performances, skill and accuracy in operational oceanography: new challenges and approaches, in: New Frontiers in Operational Oceanography, edited by: Chassignet, E., Pascual, A., Tintoré, J., and Verron, J., GODAE OceanView, 759–796, https://doi.org/10.17125/gov2018.ch29, 2018.
Hood, R. R., Laws, E. A., Armstrong, R. A., Bates, N. R., Brown, C. W., Carlson, C. A., Chai, F., Doney, S. C., Falkowski, P. G., Feely, R. A., and Friedrichs, M. A.: Pelagic functional group modeling: Progress, challenges and prospects, Deep-Sea Res. Pt. II, 53, 459–512, https://doi.org/10.1016/j.dsr2.2006.01.025, 2006.
Irby, I. D. and Friedrichs, M. A. M.: Evaluating confidence in the impact of regulatory nutrient reduction on Chesapeake Bay water quality, Estuar. Coasts, 42, 16–32, https://www.jstor.org/stable/48703007 (last access: 18 April 2025), 2019.
Irby, I. D., Friedrichs, M. A. M., Da, F., and Hinson, K. E.: The competing impacts of climate change and nutrient reductions on dissolved oxygen in Chesapeake Bay, Biogeosciences, 15, 2649–2668, https://doi.org/10.5194/bg-15-2649-2018, 2018.
Jones, E. M., Baird, M. E., Mongin, M., Parslow, J., Skerratt, J., Lovell, J., Margvelashvili, N., Matear, R. J., Wild-Allen, K., Robson, B., Rizwi, F., Oke, P., King, E., Schroeder, T., Steven, A., and Taylor, J.: Use of remote-sensing reflectance to constrain a data assimilating marine biogeochemical model of the Great Barrier Reef, Biogeosciences, 13, 6441–6469, https://doi.org/10.5194/bg-13-6441-2016, 2016.
Kishi, M. J., Kashiwai, M., Ware, D. M., Megrey, B. A., Eslinger, D. L., and Werner, F. E.: NEMURO A lower trophic level model for the north pacific marine ecosystem, Ecol. Model., 202, 12–25, https://doi.org/10.1016/j.ecolmodel.2006.08.021, 2007.
Kishi, M. J., Ito, S. I., Megrey, B. A., Rose, K. A., and Werner, F. E.: A review of the NEMURO and NEMURO.FISH models and their application to marine ecosystem investigations, J. Oceanogr., 67, 3–16, https://doi.org/10.1007/s10872-011-0009-4, 2011.
Klausmeier, C. A., Litchman, E., and Levin, S. A.: Phytoplankton growth and stoichiometry under multiple nutrient limitation, Limnol. Oceanogr., 49, 1463–1470, https://doi.org/10.4319/lo.2004.49.4_part_2.1463, 2004.
Kwiatkowski, L., Yool, A., Allen, J. I., Anderson, T. R., Barciela, R., Buitenhuis, E. T., Butenschön, M., Enright, C., Halloran, P. R., Le Quéré, C., de Mora, L., Racault, M.-F., Sinha, B., Totterdell, I. J., and Cox, P. M.: iMarNet: an ocean biogeochemistry model intercomparison project within a common physical ocean modelling framework, Biogeosciences, 11, 7291–7304, https://doi.org/10.5194/bg-11-7291-2014, 2014.
Kwiatkowski, L., Aumont, O., Bopp, L., and Ciais, P.: The impact of variable phytoplankton stoichiometry on projections of primary production, food quality, and carbon uptake in the global ocean, Global Biogeochem. Cy., 32, 516–528, 2018.
Laurent, A., Fennel, K., Cai, W.-J. Huang, W.-J, Barbero, L., and Wanninkhof, R.: Eutrophication induced acidification of coastal waters in the northern Gulf of Mexico: Insights into origin and processes from a coupled physical-biogeochemical model, Geophys. Res. Lett., 44, 946–956, https://doi.org/10.1002/2016GL071881, 2017.
Lazzari, P., Salon, S., Terzić, E., Gregg, W. W., D'Ortenzio, F., Vellucci, V., Organelli, E., and Antoine, D.: Assessment of the spectral downward irradiance at the surface of the Mediterranean Sea using the radiative Ocean-Atmosphere Spectral Irradiance Model (OASIM), Ocean Sci., 17, 675–697, https://doi.org/10.5194/os-17-675-2021, 2021.
Legendre, L. and Rassoulzadegan, F.: Plankton and nutrient dynamics in marine water, Ophelia, 41, 153–172, https://doi.org/10.1080/00785236.1995.10422042, 1995.
Lenton, T. M. and Watson, A. J.: Redfield revisited: 1. Regulation of nitrate, phosphate, and oxygen in the ocean, Global Biogeochem. Cy., 14, 225–248, https://doi.org/10.1029/1999GB900065, 2000.
Le Traon, P.-Y., Reppucci, A., Alvarez Fanjul, E., Aouf, L., Behrens, A., Belmonte, M., Bentamy, A., Bertino, L., Brando, V. E., Kreiner, M. B., Benkiran, M., Carval, T., Ciliberti, S. A., Claustre, H., Clementi, E., Coppini, G., Cossarini, G., De Alfonso Alonso-Muñoyerro, M., Delamarche, A., Dibarboure, G., Dinessen, F., Drevillon, M., Drillet, Y., Faugere, Y., Fernández, V., Fleming, A., Garcia-Hermosa, M. I., Sotillo, M. G., Garric, G., Gasparin, F., Giordan,C., Gehlen, M., Gregoire, M. L., Guinehut, S., Hamon, M., Harris, C., Hernandez, F., Hinkler, J. B., Hoyer, J., Karvonen, J., Kay, S., King, R., Lavergne, T., Lemieux-Dudon, B., Lima, L., Mao, C., Martin, M. J., Masina, S., Melet, A., Buongiorno Nardelli, B., Nolan, G., Pascual, A., Pistoia, J., Palazov, A., Piolle, J. F., Pujol, M. I., Pequignet, A. C., Peneva, E., Pérez Gómez, B., Petit de la Villeon, L., Pinardi, N., Pisano, A., Pouliquen, S., Reid, R., Remy, E., Santoleri, R., Siddorn, J., She, J., Staneva, J., Stoffelen, A., Tonani, M., Vandenbulcke, L., von Schuckmann, K., Volpe, G., Wettre, C., and Zacharioudaki, A.: From Observation to Information and Users: The Copernicus Marine Service Perspective, Front. Mar. Sci., 6, 1–22, https://doi.org/10.3389/fmars.2019.00234, 2019.
Libralato, S.: Numerical Models for Monitoring and Forecasting Ocean Ecosystems: a short description of present status, in: Ocean prediction: present status and state of the art (OPSR), edited by: Álvarez Fanjul, E., Ciliberti, S. A., Pearlman, J., Wilmer-Becker, K., and Behera, S., Copernicus Publications, State Planet, 5-opsr, 13, https://doi.org/10.5194/sp-5-opsr-13-2025, 2025.
Litchman, E. and Klausmeier, C. A.: Trait-based community ecology of phytoplankton, Annu. Rev. Ecol. Evol. S., 39, 615–639, https://doi.org/10.1146/annurev.ecolsys.39.110707.173549, 2008.
Liu, Y., Meier, H. E. M., and Eilola, K.: Nutrient transports in the Baltic Sea – results from a 30-year physical–biogeochemical reanalysis, Biogeosciences, 14, 2113–2131, https://doi.org/10.5194/bg-14-2113-2017, 2017.
Melaku Canu, D., Rosati, G., Solidoro, C., Heimbürger, L.-E., and Acquavita, A.: A comprehensive assessment of the mercury budget in the Marano–Grado Lagoon (Adriatic Sea) using a combined observational modeling approach, Mar. Chem., 177, 742–752, https://doi.org/10.1016/j.marchem.2015.10.013, 2015.
Mignot, A., Claustre, H., Cossarini, G., D'Ortenzio, F., Gutknecht, E., Lamouroux, J., Lazzari, P., Perruche, C., Salon, S., Sauzède, R., Taillandier, V., and Teruzzi, A.: Using machine learning and Biogeochemical-Argo (BGC-Argo) floats to assess biogeochemical models and optimize observing system design, Biogeosciences, 20, 1405–1422, https://doi.org/10.5194/bg-20-1405-2023, 2023.
Mitra, A., Castellani, C., Gentleman, W. C., Jónasdóttir, S. H., Flynn, K. J., Bode, A., Halsband, C., Kuhn, P., Licandro, P., Agersted, M.D., and Calbet, A.: Bridging the gap between marine biogeochemical and fisheries sciences; configuring the zooplankton link, Prog. Oceanogr., 129, 176–199, https://doi.org/10.1016/j.pocean.2014.04.025, 2014.
Mongin, M., Baird, M. E., Tilbrook, B., Matear, R. J., Lenton, A., Herzfeld, M., Wild-Allen, K., Skerratt, J., Margvelashvili, N., Robson, B. J., and Duarte, C. M.: The exposure of the Great Barrier Reef to ocean acidification, Nat. Commun., 7, 10732, https://doi.org/10.1038/ncomms10732, 2016.
Nerger, L. and Gregg, W. W.: Improving assimilation of SeaWiFS data by the application of bias correction with a local SEIK filter, J. Marine Syst., 73, 87–102, https://doi.org/10.1016/j.jmarsys.2007.09.007, 2008.
Neumann, T.: Towards a 3D-ecosystem model of the Baltic Sea, J. Marine Syst., 25, 405–419, https://doi.org/10.1016/S0924-7963(00)00030-0, 2000.
Neumann, T., Siegel, H., and Gerth, M.: A new radiation model for Baltic Sea ecosystem modelling, J. Marine Syst., 152, 83–91, https://doi.org/10.1016/j.jmarsys.2015.08.001, 2015.
Palmer, J. R. and Totterdell, I. J.: Production and export in a global ocean ecosystem model, Deep-Sea Res. Pt. I, 48, 1169–1198, https://doi.org/10.1016/S0967-0637(00)00080-7, 2001.
Pradhan, H. K., Völker, C., Losa, S. N., Bracher, A., and Nerger, L.: Global assimilation of ocean-color data of phytoplankton functional types: Impact of different data sets, J. Geophys. Res.-Oceans, 125, e2019JC015586, https://doi.org/10.1029/2019JC015586, 2020.
Redfield, A. C.: On the proportions of organic derivatives in sea water and their relation to the composition of plankton, vol. 1, University Press of Liverpool, 1934.
Rosati, G., Canu, D., Lazzari, P., and Solidoro, C.: Assessing the spatial and temporal variability of methylmercury biogeochemistry and bioaccumulation in the Mediterranean Sea with a coupled 3D model, Biogeosciences, 19, 3663–3682, https://doi.org/10.5194/bg-19-3663-2022, 2022.
Salon, S., Cossarini, G., Bolzon, G., Feudale, L., Lazzari, P., Teruzzi, A., Solidoro, C., and Crise, A.: Novel metrics based on Biogeochemical Argo data to improve the model uncertainty evaluation of the CMEMS Mediterranean marine ecosystem forecasts, Ocean Sci., 15, 997–1022, https://doi.org/10.5194/os-15-997-2019, 2019.
Schmidtko, S., Stramma, L., and Visbeck, M.: Decline in global oceanic oxygen content during the past five decades, Nature, 542, 335–339, https://doi.org/10.1038/nature21399, 2017.
Séférian, R., Berthet, S., Yool, A., Palmiéri, J., Bopp, L., Tagliabue, A., Kwiatkowski, L., Aumont, O., Christian, J., Dunne, J., and Gehlen, M.: Tracking improvement in simulated marine biogeochemistry between CMIP5 and CMIP6, Current Climate Change Reports, 6, 95–119, https://doi.org/10.1007/s40641-020-00160-0, 2020.
Skákala, J., Ford, D., Brewin, R. J., McEwan, R., Kay, S., Taylor, B., De Mora, L., and Ciavatta, S.: The assimilation of phytoplankton functional types for operational forecasting in the northwest European shelf, J. Geophys. Res.-Oceans, 123, 5230–5247, 2018.
Skákala, J., Bruggeman, J., Brewin, R. J., Ford, D. A., and Ciavatta, S.: Improved representation of underwater light field and its impact on ecosystem dynamics: A study in the North Sea, J. Geophys. Res.-Oceans, 125, e2020JC016122, https://doi.org/10.1029/2020JC016122, 2020.
Skákala, J., Ford, D., Bruggeman, J., Hull, T., Kaiser, J., King, R. R., Loveday, B., Palmer, M. R., Smyth, T., Williams, C. A., and Ciavatta, S.: Towards a multi-platform assimilative system for North Sea biogeochemistry, J. Geophys. Res.-Oceans, 126, e2020JC016649, https://doi.org/10.1029/2020JC016649, 2021.
Soetaert, K., Middelburg, J. J., Herman, P. M., and Buis, K.: On the coupling of benthic and pelagic biogeochemical models, Earth-Sci. Rev., 51, 173–201, https://doi.org/10.1016/S0012-8252(00)00004-0, 2000.
Tagliabue, A., Bopp, L., and Gehlen, M.: The response of marine carbon and nutrient cycles to ocean acidification: large uncertainties related to phytoplankton physiological assumptions, Global Biogeochem. Cy., 25, GB3017, https://doi.org/10.1029/2010GB003929, 2011.
Teruzzi, A., Dobricic, S., Solidoro, C., and Cossarini, G.: A 3-D variational assimilation scheme in coupled transport-biogeochemical models: Forecast of Mediterranean biogeochemical properties, J. Geophys. Res.-Oceans, 119, 200–217, https://doi.org/10.1002/2013JC009277, 2014.
Teruzzi, A., Bolzon, G., Feudale, L., and Cossarini, G.: Deep chlorophyll maximum and nutricline in the Mediterranean Sea: emerging properties from a multi-platform assimilated biogeochemical model experiment, Biogeosciences, 18, 6147–6166, https://doi.org/10.5194/bg-18-6147-2021, 2021.
Travers, M., Shin, Y. J., Jennings, S., Machu, E., Huggett, J. A., Field, J. G., and Cury, P. M.: Two-way coupling versus one-way forcing of plankton and fish models to predict ecosystem changes in the Benguela, Ecol. Model., 220, 3089–3099, https://doi.org/10.1016/j.ecolmodel.2009.08.016, 2009.
Verdy, A. and Mazloff, M. R.: A data assimilating model for estimating Southern Ocean biogeochemistry, J. Geophys. Res.-Oceans, 122, 6968–6988, 2017.
Vichi, M., Pinardi, N., and Masina, S.: A generalized model of pelagic biogeochemistry for the global ocean ecosystem. Part I: Theory, J. Marine Syst., 64, 89–109, https://doi.org/10.1016/j.jmarsys.2006.03.006, 2017.
Wagner, C. C., Amos, H. M., Thackray, C. P., Zhang, Y., Lundgren, E. W., Forget, G., and Sunderland, E. M.: A global 3-D ocean model for PCBs: Benchmark compounds for understanding the impacts of global change on neutral persistent organic pollutants, Global Biogeochem. Cy., 33, 469–481, https://doi.org/10.1029/2018GB006018, 2019.
Wang, B., Fennel, K., Yu, L., and Gordon, C.: Assessing the value of biogeochemical Argo profiles versus ocean color observations for biogeochemical model optimization in the Gulf of Mexico, Biogeosciences, 17, 4059–4074, https://doi.org/10.5194/bg-17-4059-2020, 2020.
Xiao, Y. and Friedrichs, M. A. M.: Using biogeochemical data assimilation to assess the relative skill of multiple ecosystem models in the Mid-Atlantic Bight: effects of increasing the complexity of the planktonic food web, Biogeosciences, 11, 3015–3030, https://doi.org/10.5194/bg-11-3015-2014, 2014.
Yumruktepe, V. Ç., Samuelsen, A., and Daewel, U.: ECOSMO II(CHL): a marine biogeochemical model for the North Atlantic and the Arctic, Geosci. Model Dev., 15, 3901–3921, https://doi.org/10.5194/gmd-15-3901-2022, 2022.
Yumruktepe, V. Ç., Mousing, E. A., Tjiputra, J., and Samuelsen, A.: An along-track Biogeochemical Argo modelling framework: a case study of model improvements for the Nordic seas, Geosci. Model Dev., 16, 6875–6897, https://doi.org/10.5194/gmd-16-6875-2023, 2023.
Zeebe, R. E. and Wolf-Gladrow, D.: CO2 in seawater: equilibrium, kinetics, isotopes, Elsevier Science, ISBN 9780444509468, 2001.
Zhang, Y., Soerensen, A. L., Schartup, A. T., and Sunderland, E. M.: A global model for methylmercury formation and uptake at the base of marine food webs, Global Biogeochem. Cy., 34, 1–21, https://doi.org/10.1029/2019GB006348, 2020.



